So this time I’m going to implement gradient descent for multivariate linear regression, but also using feature scaling. I’m using teh dataset provided in the machine learning course, which describes the cost of houses based on two parameters: the size in square feet, and the number of rooms, and giving prices in dollars.
First I’ll load the data and take a look at it.
library(dplyr)
library(magrittr)
library(ggplot2)
"ex1data2.txt" %>%
read.csv(
header = FALSE,
col.names = c("size","n_rooms","price")
) %>%
dplyr::mutate(
n_rooms = factor(n_rooms)
) -> house_prices
house_prices %>% head## size n_rooms price
## 1 2104 3 399900
## 2 1600 3 329900
## 3 2400 3 369000
## 4 1416 2 232000
## 5 3000 4 539900
## 6 1985 4 299900So we have two $x$’s: size and n_rooms
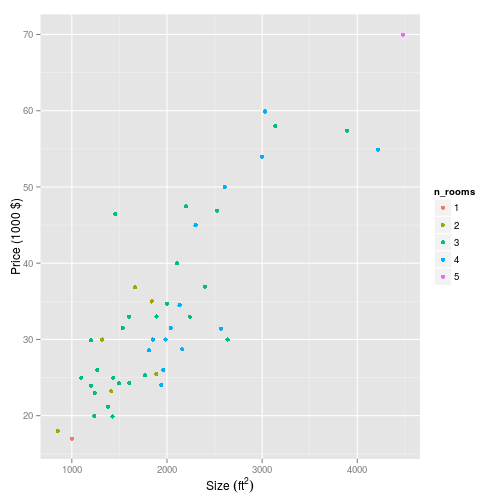
Let’s also plot it out of interest:
p <- house_prices %>%
ggplot(
aes(
x = size,
y = price,
colour = n_rooms
)
) +
geom_point()+
scale_x_continuous(expression(Size~(ft^2)))+
scale_y_continuous(
"Price (1000 $)",
breaks = seq(2e+05,7e+05,1e+05),
labels = seq(20,70,10)
)
pFeature normalisation/scaling
To copy the exercise document:
Your task here is to complete the code in featureNormalize.m to
- Subtract the mean value of each feature from the dataset.
- After subtracting the mean, additionally scale (divide) the feature values by their respective “standard deviations.”
and in the file featureNormalize.m provided with the course material, we get:
First, for each feature dimension, compute the mean of the feature and subtract it from the dataset, storing the mean value in mu. Next, compute the standard deviation of each feature and divide each feature by it’s standard deviation, storing the standard deviation in sigma.
Note that X is a matrix where each column is a feature and each row is an example. You need to perform the normalization separately for each feature.
I’ll have a go at implementing that in R.
feature_scale <- function(x) {
# Convert all factors to numeric
# Note that this will also allow the conversion of string features
for (i in 1:ncol(x)) {
x[,i] %>% as.numeric -> x[,i]
}
# Set up matrices to take outputs
mu <- matrix(nrow = 1, ncol = ncol(x))
sigma <- matrix(nrow = 1, ncol = ncol(x))
scaled <- matrix(nrow = nrow(x), ncol = ncol(x))
# Define feature scaling function
scale <- function(feature) {
(feature - mean(feature)) / sd(feature)
}
# Run this for each of the features
for (i in 1:ncol(x)) {
mu[,i] <- mean(x[,i])
sigma[,i] <- sd(x[,i])
scaled[,i] <- scale(x[,i])
}
# And output them together as a list
list(
mu = mu,
sigma = sigma,
scaled = scaled
)
}Ok so let’s try this on our features in the housing dataset.
scaled_features <- feature_scale(house_prices[,-3])We can have a look to see what this has done to our values. Originally the ranges for the features were:
house_prices %$% size %>% range## [1] 852 4478and
house_prices %$% n_rooms %>% as.character %>% as.numeric %>% range ## [1] 1 5…so quite a difference.
After feature scaling these ranges are:
scaled_features %$% scaled %>% extract(,1) %>% range## [1] -1.445423 3.117292and
scaled_features %$% scaled %>% extract(,2) %>% range ## [1] -2.851859 2.404508…so now much closer.
Gradient descent
In the multivariate case, the cost function can also be written in the vectorised form:
Where:
grad <- function(alpha, j, X, y, theta) {
# J <- function(X, y, theta) {
# sum( (X %*% theta - y)^2 ) / (2*length(y))
# }
# The cost function vectorises to:
J <- function(X, y, theta) {
(1/2*length(y)) * t((X %*% theta - y)) %*% (X %*% theta - y)
}
theta_history <<- matrix(nrow = j, ncol = ncol(X) + 1)
for (i in 1:j) {
error <- (X %*% theta - y)
delta <- t(X) %*% error / length(y)
theta <- theta - alpha * delta
theta_history[i,] <<- c(theta,J(X, y, theta))
if (i > 1) {
# Here I define a function to calculate when we have roughly reached convergence.
if (
isTRUE(
all.equal(
theta_history[i,3],
theta_history[i-1,3]
#tolerance = # can set a tolerance here if required.
)
)
) {
theta_history <<- theta_history[1:i,]
break
}
}
}
list(
theta = theta,
cost = theta_history[i,3],
iterations = i
)
}Here I use the grad() gradient descent function I defined in my post about linear regression with gradient descent.
First set up the inputs:
X <- matrix(ncol = ncol(house_prices)-1,nrow = nrow(house_prices))
X[,1:2] <- cbind(house_prices$size, house_prices$n_rooms)
X <- cbind(1, X)
y <- matrix(house_prices$price, ncol = 1)
theta <- matrix(rep(0,3), ncol = 1)And simply apply the function, but on the raw data without feature scaling.
multi_lin_reg <- grad(
alpha = 0.1,
j = 1000,
X = X,
y = y,
theta = theta
) %>% print## $theta
## [,1]
## [1,] NaN
## [2,] NaN
## [3,] NaN
##
## $cost
## [1] NaN
##
## $iterations
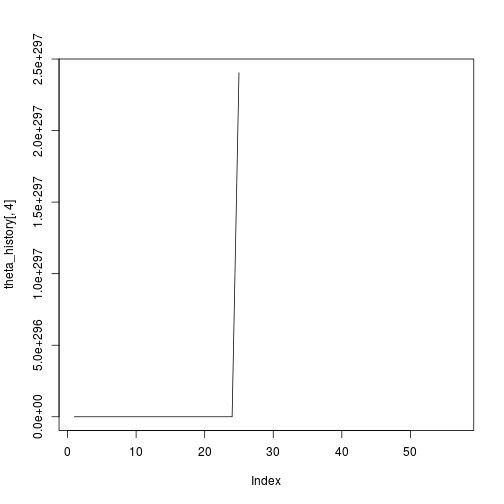
## [1] 57Hmm ok so that didn’t seem to work. Just out of interest, let’s plot the history:
plot(theta_history[,4],type="l")Definitely something not working there. Ok so now I’ll try it with feature scaling.
X[,2:3] <- feature_scale(X[,2:3])[[3]]
multi_lin_reg <- grad(
alpha = 0.1,
j = 1000,
X = X,
y = y,
theta = theta
) %>% print## $theta
## [,1]
## [1,] 340412.660
## [2,] 110631.048
## [3,] -6649.472
##
## $cost
## [1] -6649.472
##
## $iterations
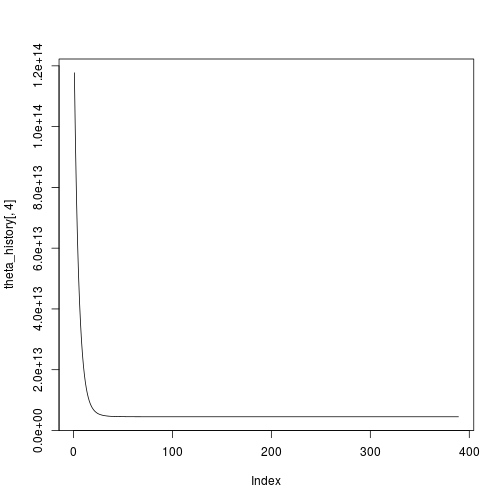
## [1] 389And to plot it:
plot(theta_history[,4],type="l")Great, convergence after 389 iterations. All seems well, but I want to compare this with a multiple linear regression the traditional way:
model <- lm(
price ~ size + n_rooms,
data = house_prices %>% mutate(n_rooms = as.integer(n_rooms))
)
coef(model)## (Intercept) size n_rooms
## 89597.9095 139.2107 -8738.0191The parameters don’t match, but this is because we have scaled the features. The output from the two models will be the same. Here I check by combining the two predictions into the house_prices dataframe, and comparing them with identical().
house_prices %<>%
dplyr::mutate(
# Vectorised method of theta transpose X
vector_pred = (X %*% multi_lin_reg$theta),
# Traditional statistical method of y = a + bx + cx
pred = coef(model)[1] + (coef(model)[2] * size) + (coef(model)[3]*as.integer(n_rooms))
)
identical(
c(house_prices$vector_pred),
c(house_prices$pred)
)## [1] FALSEOk not identical, how come?
(house_prices$pred - house_prices$vector_pred) %>% mean## [1] 3.244767e-10So they differ by a pretty small amount. Try the comparison more sensibly:
all.equal(
c(house_prices$vector_pred),
c(house_prices$pred)
)## [1] TRUEAnd now let’s plot the actual data with predictions from the multiple regression.
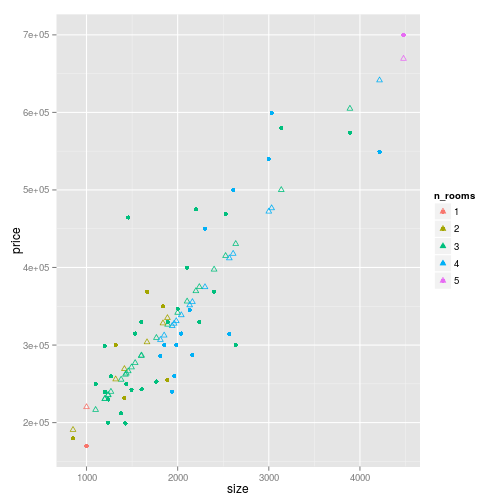
house_prices %>%
ggplot(
aes(
x = size,
y = price,
colour = n_rooms
)
) +
geom_point() +
geom_point(
aes(
x = size,
y = vector_pred
),
shape = 2,
)Pretty close to a single regression model, but you can see that there are slightly different slopes for each number of rooms.